seaborn.PairGrid(**kwargs)¶Subplot grid for plotting pairwise relationships in a dataset.
This object maps each variable in a dataset onto a column and row in a grid of multiple axes. Different axes-level plotting functions can be used to draw bivariate plots in the upper and lower triangles, and the the marginal distribution of each variable can be shown on the diagonal.
Several different common plots can be generated in a single line using
pairplot(). Use PairGrid when you need more flexibility.
See the tutorial for more information.
__init__(self, data, *, hue=None, hue_order=None, palette=None, hue_kws=None, vars=None, x_vars=None, y_vars=None, corner=False, diag_sharey=True, height=2.5, aspect=1, layout_pad=0.5, despine=True, dropna=False, size=None)¶Initialize the plot figure and PairGrid object.
Tidy (long-form) dataframe where each column is a variable and each row is an observation.
Variable in data to map plot aspects to different colors. This
variable will be excluded from the default x and y variables.
Order for the levels of the hue variable in the palette
Set of colors for mapping the hue variable. If a dict, keys
should be values in the hue variable.
Other keyword arguments to insert into the plotting call to let other plot attributes vary across levels of the hue variable (e.g. the markers in a scatterplot).
Variables within data to use, otherwise use every column with
a numeric datatype.
Variables within data to use separately for the rows and
columns of the figure; i.e. to make a non-square plot.
If True, don’t add axes to the upper (off-diagonal) triangle of the grid, making this a “corner” plot.
Height (in inches) of each facet.
Aspect * height gives the width (in inches) of each facet.
Padding between axes; passed to fig.tight_layout.
Remove the top and right spines from the plots.
Drop missing values from the data before plotting.
See also
Examples
Calling the constructor sets up a blank grid of subplots with each row and one column corresponding to a numeric variable in the dataset:
penguins = sns.load_dataset("penguins")
g = sns.PairGrid(penguins)
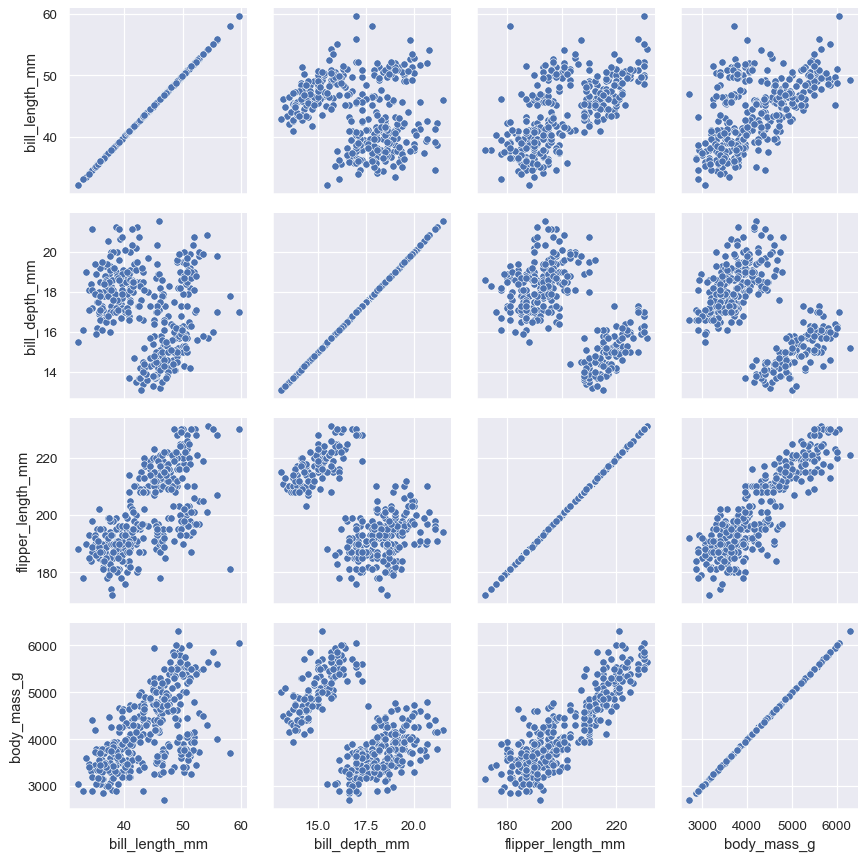
Passing a bivariate function to PairGrid.map() will draw a bivariate plot on every axes:
g = sns.PairGrid(penguins)
g.map(sns.scatterplot)
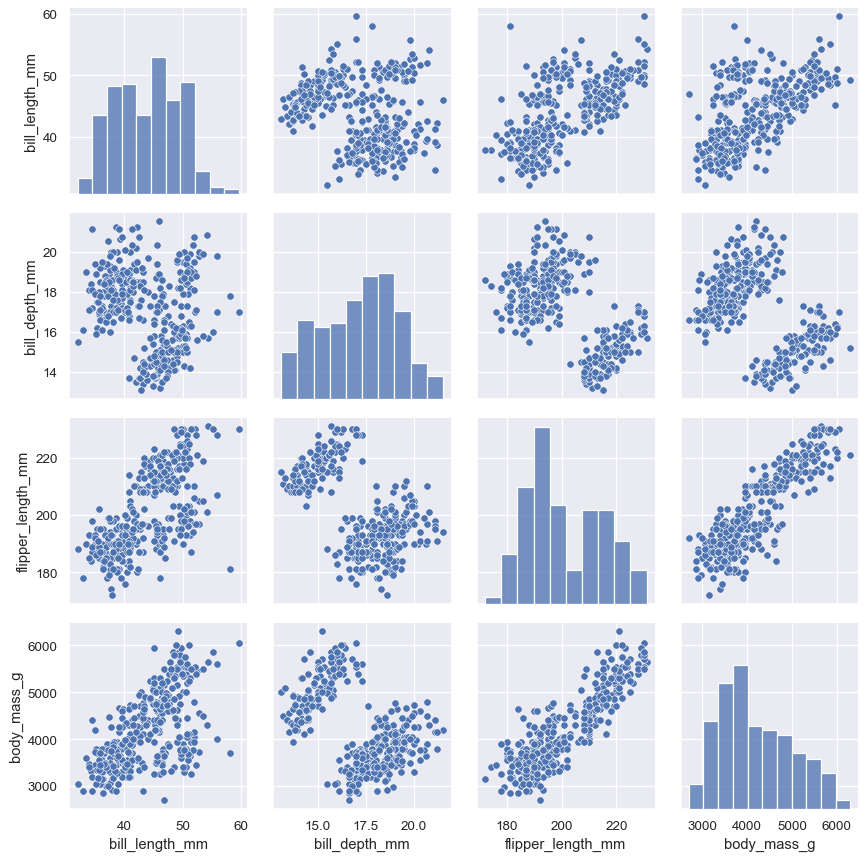
Passing separate functions to PairGrid.map_diag() and PairGrid.map_offdiag() will show each variable’s marginal distribution on the diagonal:
g = sns.PairGrid(penguins)
g.map_diag(sns.histplot)
g.map_offdiag(sns.scatterplot)
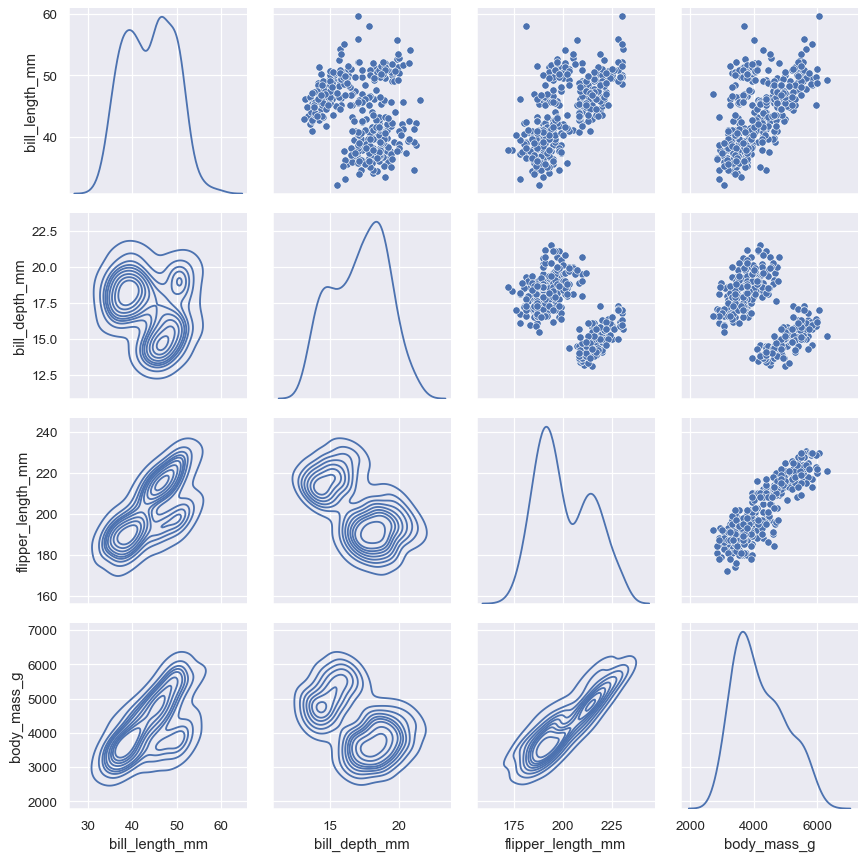
It’s also possible to use different functions on the upper and lower triangles of the plot (which are otherwise redundant):
g = sns.PairGrid(penguins, diag_sharey=False)
g.map_upper(sns.scatterplot)
g.map_lower(sns.kdeplot)
g.map_diag(sns.kdeplot)
Or to avoid the redundancy altogether:
g = sns.PairGrid(penguins, diag_sharey=False, corner=True)
g.map_lower(sns.scatterplot)
g.map_diag(sns.kdeplot)
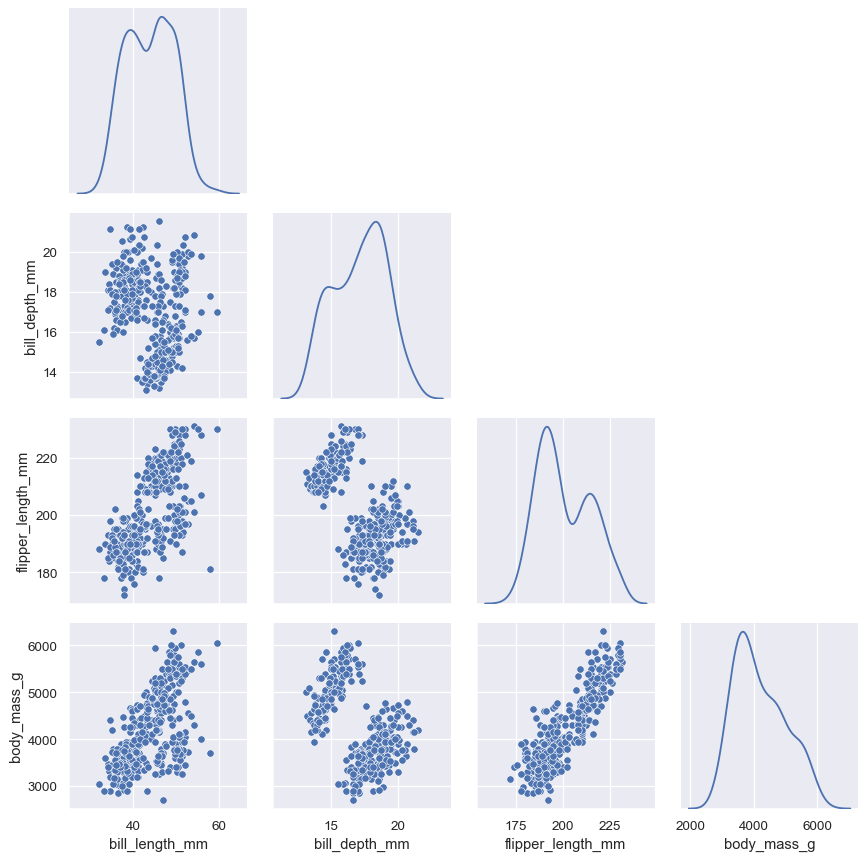
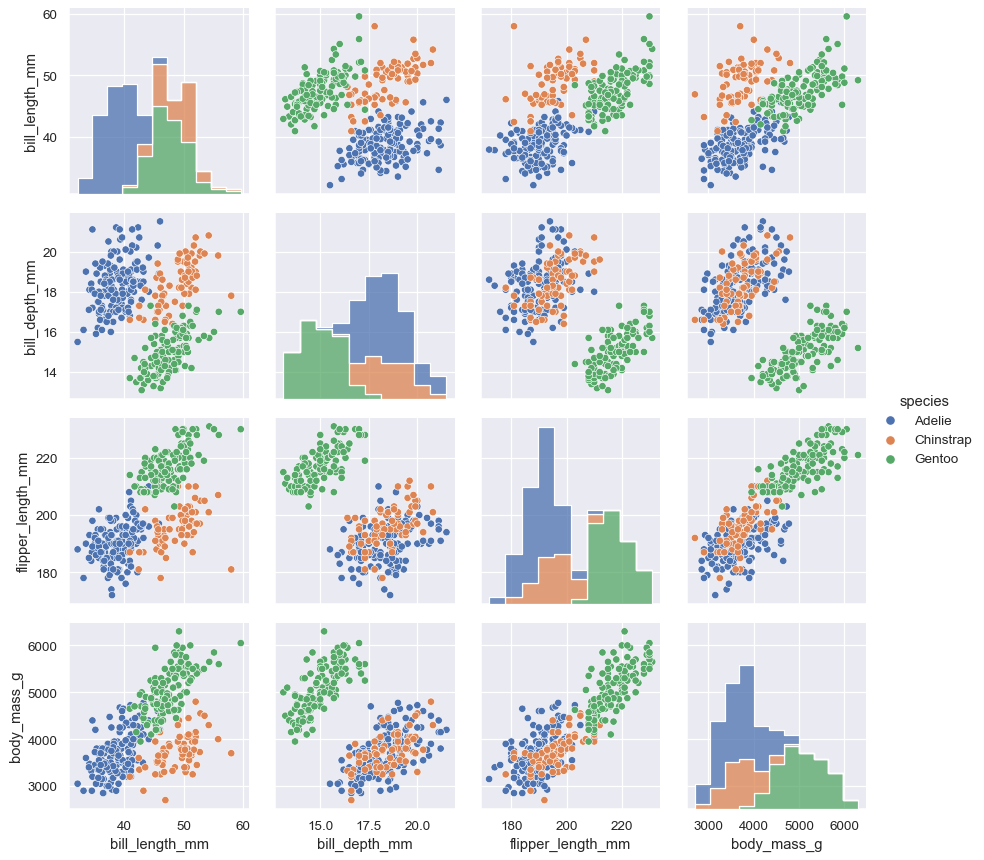
The PairGrid constructor accepts a hue variable. This variable is passed directly to functions that understand it:
g = sns.PairGrid(penguins, hue="species")
g.map_diag(sns.histplot)
g.map_offdiag(sns.scatterplot)
g.add_legend()
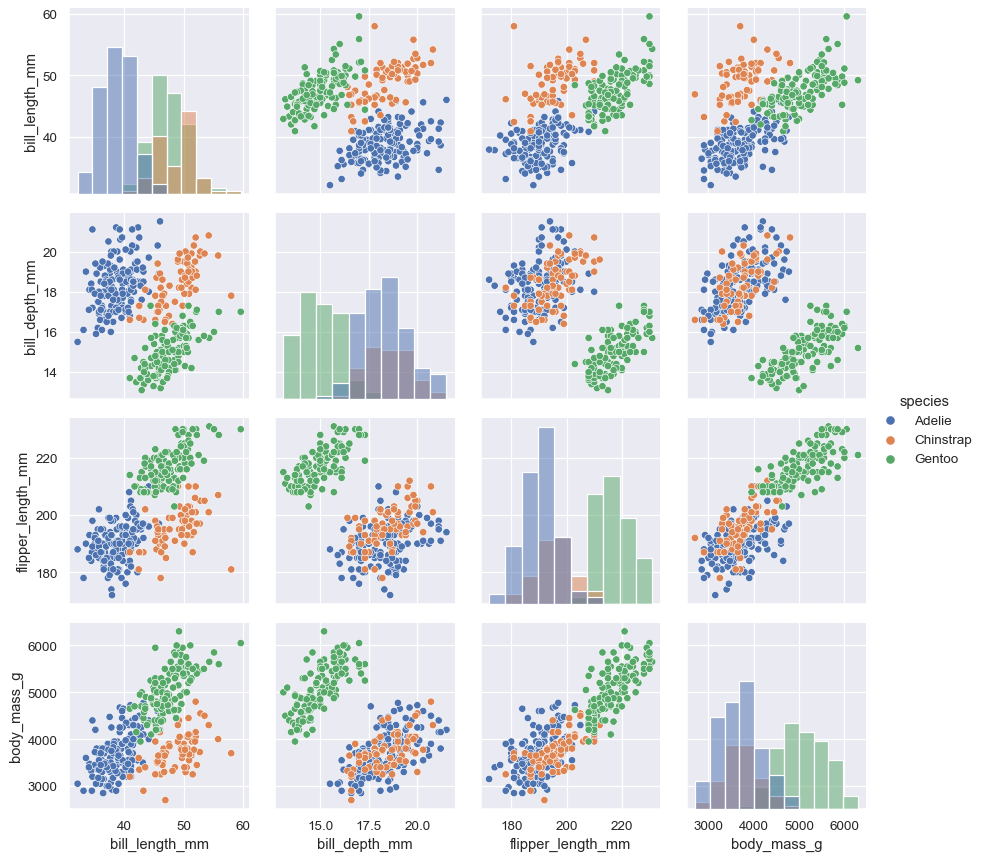
But you can also pass matplotlib functions, in which case a groupby is performed internally and a separate plot is drawn for each level:
g = sns.PairGrid(penguins, hue="species")
g.map_diag(plt.hist)
g.map_offdiag(plt.scatter)
g.add_legend()
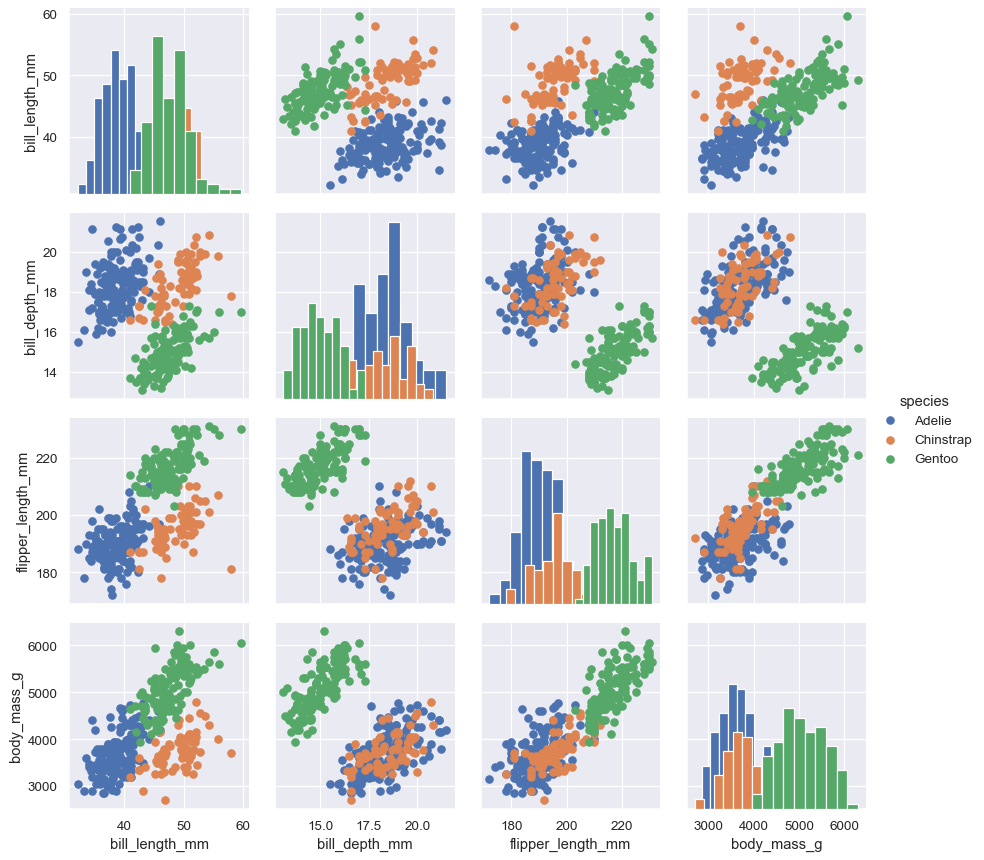
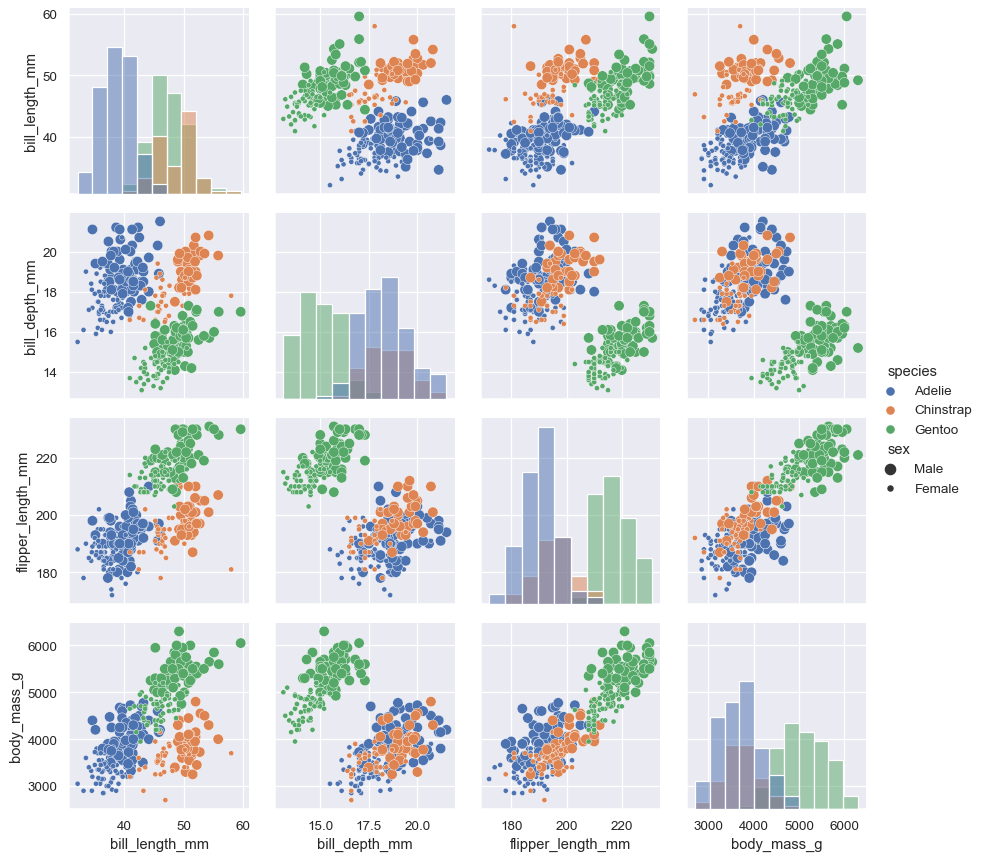
Additional semantic variables can be assigned by passing data vectors directly while mapping the function:
g = sns.PairGrid(penguins, hue="species")
g.map_diag(sns.histplot)
g.map_offdiag(sns.scatterplot, size=penguins["sex"])
g.add_legend(title="", adjust_subtitles=True)
When using seaborn functions that can implement a numeric hue mapping, you will want to disable mapping of the variable on the diagonal axes. Note that the hue variable is excluded from the list of variables shown by default:
g = sns.PairGrid(penguins, hue="body_mass_g")
g.map_diag(sns.histplot, hue=None, color=".3")
g.map_offdiag(sns.scatterplot)
g.add_legend()
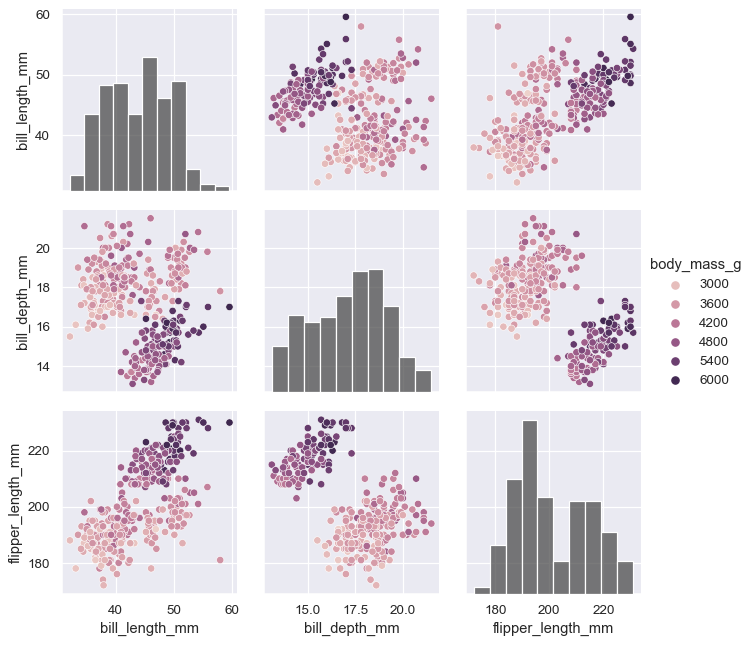
The vars parameter can be used to control exactly which variables are used:
variables = ["body_mass_g", "bill_length_mm", "flipper_length_mm"]
g = sns.PairGrid(penguins, hue="body_mass_g", vars=variables)
g.map_diag(sns.histplot, hue=None, color=".3")
g.map_offdiag(sns.scatterplot)
g.add_legend()
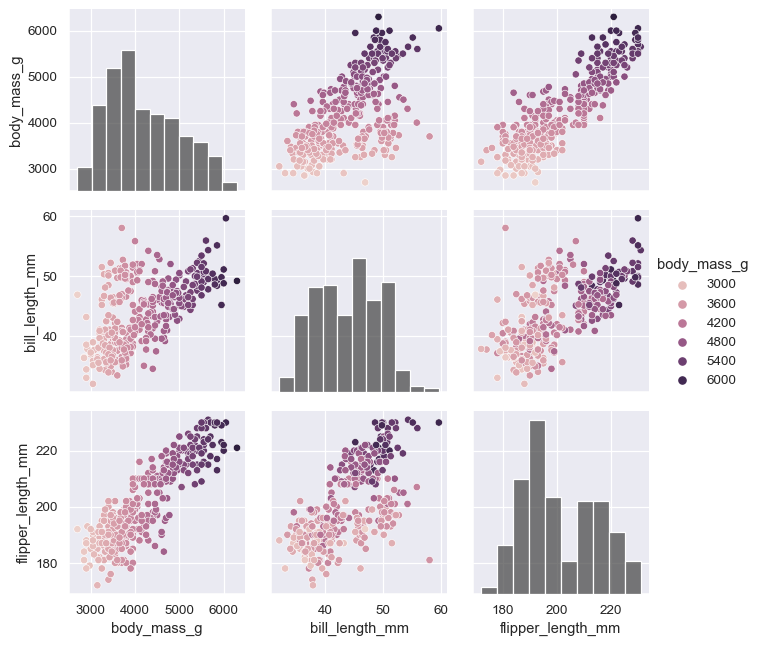
The plot need not be square: separate variables can be used to define the rows and columns:
x_vars = ["body_mass_g", "bill_length_mm", "bill_depth_mm", "flipper_length_mm"]
y_vars = ["body_mass_g"]
g = sns.PairGrid(penguins, hue="species", x_vars=x_vars, y_vars=y_vars)
g.map_diag(sns.histplot, color=".3")
g.map_offdiag(sns.scatterplot)
g.add_legend()
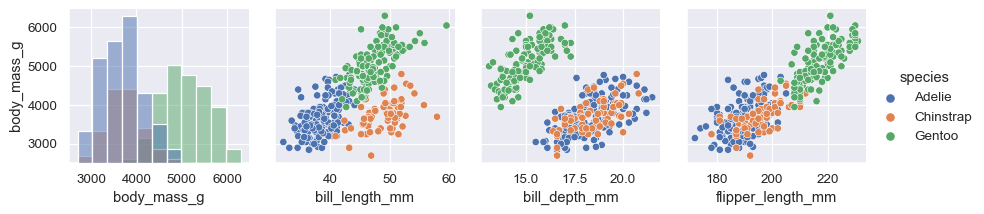
It can be useful to explore different approaches to resolving multiple distributions on the diagonal axes:
g = sns.PairGrid(penguins, hue="species")
g.map_diag(sns.histplot, multiple="stack", element="step")
g.map_offdiag(sns.scatterplot)
g.add_legend()
Methods
|
Initialize the plot figure and PairGrid object. |
|
Draw a legend, maybe placing it outside axes and resizing the figure. |
|
Plot with the same function in every subplot. |
|
Plot with a univariate function on each diagonal subplot. |
|
Plot with a bivariate function on the lower diagonal subplots. |
|
Plot with a bivariate function on the off-diagonal subplots. |
|
Plot with a bivariate function on the upper diagonal subplots. |
|
Save an image of the plot. |
|
Set attributes on each subplot Axes. |
|
Call fig.tight_layout within rect that exclude the legend. |
Attributes
|
DEPRECATED: prefer the |
|
Access the |
|
The |